stdClass Object
(
[nazev] => REPARES
[adresa_url] =>
[api_hash] =>
[seo_desc] =>
[jazyk] => en
[jednojazycny] =>
[barva] =>
[indexace] => 1
[obrazek] =>
[ga_force] =>
[cookie_force] =>
[secureredirect] => 1
[google_verification] => UOa3DCAUaJJ2C3MuUhI9eR1T9ZNzenZfHPQN4wupOE8
[ga_account] => UA-10822215-5
[ga_domain] =>
[ga4_account] => G-VKDBFLKL51
[gtm_id] =>
[gt_code] =>
[kontrola_pred] =>
[omezeni] => 0
[pozadi1] => 0003~~C8ivSkw5vFYhyDXAMcg1WKEktbgEAA.png
[pozadi2] =>
[pozadi3] =>
[pozadi4] =>
[pozadi5] =>
[robots] =>
[htmlheaders] =>
[newurl_domain] => 'repares.vscht.cz'
[newurl_jazyk] => 'en'
[newurl_akce] => '[en]'
[newurl_iduzel] =>
[newurl_path] => 8549/49296/49297
[newurl_path_link] => Odkaz na newurlCMS
[iduzel] => 49297
[platne_od] => 27.08.2024 14:12:00
[zmeneno_cas] => 27.08.2024 14:12:18.974251
[zmeneno_uzivatel_jmeno] => Jan Kříž
[canonical_url] =>
[idvazba] => 55725
[cms_time] => 1751839337
[skupina_www] => Array
(
)
[slovnik] => stdClass Object
(
[preloader] => Wait a second...
[logo_href] => /
[logo] =>  [logo_mobile_href] => /
[logo_mobile] =>
[logo_mobile_href] => /
[logo_mobile] =>  [google_search] =>
[social_in_odkaz] =>
[social_fb_odkaz] => https://www.facebook.com/utvpvscht/
[social_tw_odkaz] =>
[social_yt_odkaz] =>
[intranet_odkaz] =>
[intranet_text] =>
[mobile_over_nadpis_menu] => Menu
[mobile_over_nadpis_search] => Search
[mobile_over_nadpis_jazyky] => Languages
[mobile_over_nadpis_login] => Login
[menu_home] => Homepage
[more_info] =>
[paticka_mapa_odkaz] =>
[paticka_budova_a_nadpis] => BUILDING A
[paticka_budova_a_popis] => Rector,
Department of Communications,
Department of Education,
FCT Dean’s Office,
Centre for Information Services
[paticka_budova_b_nadpis] => BUILDING B
[paticka_budova_b_popis] => Department of R&D, Dean’s Offices:
FET,
FFBT,
FCE,
Computer Centre,
Department of International Relations,
Bursar
[paticka_budova_c_nadpis] => BUILDING C
[paticka_budova_c_popis] => Crèche Zkumavka,
General Practitioner,
Department of Economics and Management,
Department of Mathematics
[paticka_budova_1_nadpis] => NATIONAL LIBRARY OF TECHNOLOGY
[paticka_budova_1_popis] =>
[paticka_budova_2_nadpis] => CAFÉ CARBON
[paticka_budova_2_popis] =>
[paticka_adresa] => UCT Prague
[google_search] =>
[social_in_odkaz] =>
[social_fb_odkaz] => https://www.facebook.com/utvpvscht/
[social_tw_odkaz] =>
[social_yt_odkaz] =>
[intranet_odkaz] =>
[intranet_text] =>
[mobile_over_nadpis_menu] => Menu
[mobile_over_nadpis_search] => Search
[mobile_over_nadpis_jazyky] => Languages
[mobile_over_nadpis_login] => Login
[menu_home] => Homepage
[more_info] =>
[paticka_mapa_odkaz] =>
[paticka_budova_a_nadpis] => BUILDING A
[paticka_budova_a_popis] => Rector,
Department of Communications,
Department of Education,
FCT Dean’s Office,
Centre for Information Services
[paticka_budova_b_nadpis] => BUILDING B
[paticka_budova_b_popis] => Department of R&D, Dean’s Offices:
FET,
FFBT,
FCE,
Computer Centre,
Department of International Relations,
Bursar
[paticka_budova_c_nadpis] => BUILDING C
[paticka_budova_c_popis] => Crèche Zkumavka,
General Practitioner,
Department of Economics and Management,
Department of Mathematics
[paticka_budova_1_nadpis] => NATIONAL LIBRARY OF TECHNOLOGY
[paticka_budova_1_popis] =>
[paticka_budova_2_nadpis] => CAFÉ CARBON
[paticka_budova_2_popis] =>
[paticka_adresa] => UCT Prague
Technická 5
166 28 Prague 6 – Dejvice
IČO: 60461373 / VAT: CZ60461373
Czech Post certified digital mail code: sp4j9ch
Copyright: UCT Prague 2017
Information provided by the Department of International Relations and the Department of R&D. Technical support by the Computing Centre.
[paticka_odkaz_mail] => mailto:Jan.Bartacek@vscht.cz
[zobraz_desktop_verzi] => switch to desktop version
[drobecky] => You are here: UCT Prague –
[aktualizovano] => Updated
[autor] => Author
[social_in_title] =>
[social_fb_title] =>
[den_kratky_4] =>
[archiv_novinek] =>
[novinky_servis_archiv_rok] =>
[novinky_kategorie_1] =>
[novinky_kategorie_2] =>
[novinky_kategorie_3] =>
[novinky_kategorie_4] =>
[novinky_kategorie_5] =>
[novinky_archiv_url] =>
[novinky_servis_nadpis] =>
[novinky_dalsi] =>
[zobraz_mobilni_verzi] =>
[den_kratky_2] =>
[den_kratky_1] =>
[den_kratky_3] =>
[nepodporovany_prohlizec] =>
[den_kratky_5] =>
[den_kratky_0] =>
[social_li_odkaz] =>
[hledani_nadpis] => hledání
[hledani_nenalezeno] => Nenalezeno...
[hledani_vyhledat_google] => vyhledat pomocí Google
[stahnout] =>
[dokumenty_kod] =>
[dokumenty_nazev] =>
[dokumenty_platne_od] =>
[dokumenty_platne_do] =>
)
[poduzel] => stdClass Object
(
[49299] => stdClass Object
(
[obsah] =>
[poduzel] => stdClass Object
(
[49304] => stdClass Object
(
[nazev] =>
[seo_title] => REPARES
[seo_desc] =>
[autor] =>
[autor_email] =>
[obsah] =>
[urlnadstranka] =>
[ogobrazek] =>
[pozadi] =>
[iduzel] => 49304
[canonical_url] =>
[skupina_www] => Array
(
)
[url] => /home
[sablona] => stdClass Object
(
[class] => boxy
[html] =>
[css] =>
[js] => $(function() {
setInterval(function () { $('*[data-countdown]').each(function() { CountDownIt('#'+$(this).attr("id")); }); },1000);
setInterval(function () { $('.homebox_slider:not(.stop)').each(function () { slide($(this),true); }); },5000);
});
function CountDownIt(selector) {
var el=$(selector);foo = new Date;
var unixtime = el.attr('data-countdown')*1-parseInt(foo.getTime() / 1000); if(unixtime<0) unixtime=0;
var dnu = 1*parseInt(unixtime / (3600*24)); unixtime=unixtime-(dnu*(3600*24));
var hodin = 1*parseInt(unixtime / (3600)); unixtime=unixtime-(hodin*(3600));
var minut = 1*parseInt(unixtime / (60)); unixtime=unixtime-(minut*(60));
if(unixtime<10) {unixtime='0'+unixtime;}
if(dnu<10) {unixtime='0'+dnu;}
if(hodin<10) {unixtime='0'+hodin;}
if(minut<10) {unixtime='0'+minut;}
el.html(dnu+':'+hodin+':'+minut+':'+unixtime);
}
function slide(el,vlevo) {
if(el.length<1) return false; var leva=el.find('.content').position().left; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1;
var cislo=leva/sirka*-1; if(vlevo) { if(cislo+1>pocet) cislo=0; else cislo++; } else { if(cislo==0) cislo=pocet-1; else cislo--; }
el.find('.content').animate({'left':-1*cislo*sirka});
el.find('.slider_puntiky a').removeClass('selected');
el.find('.slider_puntiky a.puntik'+cislo).addClass('selected');
return false;
}
function slideTo(el,cislo) {
if(el.length<1) return false; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; if(cislo<0 || cislo>pocet) return false;
el.find('.content').animate({'left':-1*cislo*sirka});
el.find('.slider_puntiky a').removeClass('selected');
el.find('.slider_puntiky a.puntik'+cislo).addClass('selected');
return false;
}
[autonomni] => 1
)
)
[69928] => stdClass Object
(
[nazev] => Antibiotická rezistence v ČR: Jak společně zastavit nezastavitelné
[seo_title] => Antibiotická rezistence v ČR: Jak společně zastavit nezastavitelné
[seo_desc] => Pozvánka na důležitý seminář o antibiotické rezistenci (REPARES, CZEPAR, VŠCHT Praha)
[autor] => Lucie Pokorná
[autor_email] => krayzell@vscht.cz
[perex] =>
[ikona] =>
[obrazek] =>
[ogobrazek] =>
[pozadi] =>
[obsah] => Česká platforma antibiotické rezistence Vás zve na seminář:
Antibiotická rezistence v ČR: Jak společně zastavit nezastavitelné
seminář o medicíně, mikrobiologii a životním prostředí pro širší odbornou veřejnost se díky projektu REPARES konal 17.5. 2023 od 9:30 ve Smetanově sále Autoklubu ČR (Opletalova 29, Praha 1)
Boj s antibiotickou rezistencí vyžaduje multidisciplinární týmy expertů z oblasti humánní a veterinární medicíny, mikrobiologie a environmentální technologie. Cílem vznikající České platformy antibiotické rezistence (CZEPAR) je sdružovat odborníky z akademické sféry, praktické medicíny, firemního sektoru a státní správy.
Chcete se dozvědět více? Zaregistrujte se na seminář CZEPAR o možnostech mezioborového řešení problematiky antibiotické rezistence v České republice.
Program semináře
|
09:00 – 09:30 |
Registrace účastníků |
|
09:30 – 09:40 |
Úvod semináře - Ing. Dana Kok, Ph.D. / Prof. Ing. Jan Bartáček, Ph.D. (Vysoká škola chemicko-technologická v Praze) |
|
09:40 – 09:55 |
Fenomén antibiotická rezistence - doc. MUDr. Helena Žemličková, Ph.D. (Státní zdravotní ústav) |
|
10:00 – 10:15 |
- MUDr. Jan Závora a Ing. Gabriela Kroneislová (Všeobecné fakultní nemocnice a 1. Lékařská fakulta Univerzity Karlovy) |
|
10:20 – 10:35 |
Meticilin rezistentní Staphylococcus aureus v českých a slovenských nemocnicích a komunitě - Mgr. Jan Tkadlec, Ph.D. (Fakultní nemocnice Motol, 2. Lékařská fakulta UK) |
|
10:40 – 11:00 |
občerstvení |
|
11:00 – 11:20 |
Problematika antibiotické rezistence z pohledu veterinární medicíny - MVDr. Tomáš Černý (Státní veterinární ústav Praha) |
|
11:25 – 11:45 |
Interakce mezi společností a životním prostředím – One Health koncept - doc. RNDr. Monika Dolejská, Ph.D. (Ústav laboratorní medicíny, Fakultní nemocnice Brno, CEITEC) |
|
11:50 – 12:10 |
Antibiotická rezistence očima Evropské unie - MUDr. Pavel Březovský, MBA (Státní zdravotní ústav) |
|
12:10 – 13:00 |
oběd |
|
13:10 – 14:30 |
Panelová diskuse (1 moderátor, 3 – 4 panelisté) |
|
14:30 – 14:50 |
občerstvení |
|
14:50 – 15:05 |
Antibiotická rezistence na čistírnách odpadních vod - Ing. Petra Moťková, Ph.D. a Ing. Iveta Brožková, Ph.D. (Univerzita Pardubice) |
|
15:10 – 15:25 |
Antibiotická rezistence ve vodách, kalech a dopady na životní prostředí i lidské zdraví - Ing. Sabina Purkrtová, Ph.D. (Vysoká škola chemicko-technologická v Praze) |
|
15:30 – 15:45 |
Česká platforma antibiotické rezistence: Jak společně zastavit nezastavitelné - Ing. Dana Kok, Ph.D. / Prof. Ing. Jan Bartáček, Ph.D. (Vysoká škola chemicko-technologická v Praze) |
|
15:45 – 15:55 |
Shrnutí semináře a pozvání na závěrečnou diskusi u občerstvení |
|
16:00 – ??:?? |
Závěrečný networking |
REPARES (Research Platform on Antibiotic Resistance Spread through Wastewater Treatment Plants), a consortium of leading academic, industry, and policymakers knowledgeable in the Antibiotic Resistance (AR) subject, is geared towards contributing excellence in research on the spread of AR in wastewater treatment plants (WWTPs).
[ikona] => [obrazek] => 0003~~MwMA.png [ogobrazek] => [pozadi] => [obsah] =>
The REPARES Team, mostly made up of academic leaders from top universities across Europe, has established strong cooperation with leading European innovators and internationally renowned experts in the water-related antibiotic resistance field.
- UCT will coordinate and work with AR experts to provide methodologies and development of the REPARES platform
- UCP will leverage its expert leadership in wastewater-mediated antibiotic resistance.
- TUDelft will deliver methods to apply for international projects successfully.
- Wetsus will propel cooperation with the non-academic sector.
- AAU will integrate the REPARES database within the world-accepted MiDAS.
- UW will do the bioinformatics and the validation of qPCR primers.
Background
Antibiotics were developed in 1940 solely for the treatment of infections in humans and animals. Over the years, diseases considered deadly were considered non-issues due to the effectiveness of these medicines. Regrettably, the benefits of antibiotics have been hampered by the development of resistance to antibiotics by bacteria. In recent times certain antibiotics used to eradicate some bacteria have become inactive. Antibiotic resistance (AR) has become prevalent, spreading primarily in populations and the environment. Thus, the development and proliferation of antibiotic resistance are classified among major global threats by the World Health Organization ( ). In addition, the European Centre for Disease Prevention and Control described the colossal effect of AR on the human population resulting in approximately 25,000 deaths per annum, costing the health care sector over € 1.5 billion every year.
Proper sanitation of the water cycle is essential to sustain environmental and human health. The role of wastewater treatment plants (WWTPs) and their microbiomes as putative hotspots for the proliferation of antibiotic resistance has questioned how effective these treatment systems are. Increasingly rising public, stakeholders, and governmental perception on this issue has highlighted the need for new quality criteria and installation upgrades.
Despite the gravity of the AR problem, in the wastewater sector, methodological biases have been the sole limiting factor on adequately monitoring the emission, fate, and possible amplification or removal of antibiotic-resistant bacteria (ARB) and antibiotic resistance genes (ARGs) across WWTPs. Indeed, there is an urgent need to develop standardized methods and a well-curated database for accurate identification, characterization, and quantification of antibiotic resistance in such complex water matrices as wastewater. These issues are what REPARES aims to solve during the project period.
 REPARES Goals
REPARES Goals
REPARES aims at advancing the European community’s know-how on antibiotic resistance across sanitation waterways, which in the future will bring society closer to achieving the Sustainable Development Goal of the United Nations. REPARES will act as an important vector to disseminate information on antimicrobial resistance spread within European WWTPs beyond science and technology to reach the academic and non-academic communities utilizing open events, popularization publications, and operation of the REPARES web platform.
Duration of Project
1 October 2019 - 30 September 2022
The relevance of research on antibiotic resistance
- Antibiotics are one of the common contaminants in wastewater.
- The increased bacterial antibiotic resistance stems from the spread of antibiotic-resistant bacteria and the transfer of resistance genes.
- Antibiotic resistance can be acquired through random gene mutations and/or by horizontal gene transfer.
- The World Health Organization qualified antibiotic resistance development as a significant global threat to society.
REPARES Objectives
- The increasing level of UCT’s scientific excellence
- Improving scientific profiles of UCT’s researchers
- Increasing UCT’s ability to apply for international projects successfully
- Fostering cooperation with the non-academic sector
- Advancing European community’s know-how on antibiotic resistance occurrence and spread in Europe
In total 49 members (except for REPARES members) joined the REPARES platform. The members are in total from 20 countries all around the world. The highest amount of people is from the Czech Republic, however, there are also more people from Belgium, Columbia, Denmark, France, the Netherlands, Poland, the United States of America, and the South African Republic. We also have one member from Argentina, Canada, Egypt, Great Britain, Ireland, Italy, Mexico, Spain, Sweden, Switzerland, and Norway.
Most of the members are from universities (29 members), companies (8 members), policymakers (4 members), institutions (3), and laboratories (2 members). There is also one member from the platform and 2 undefined members.
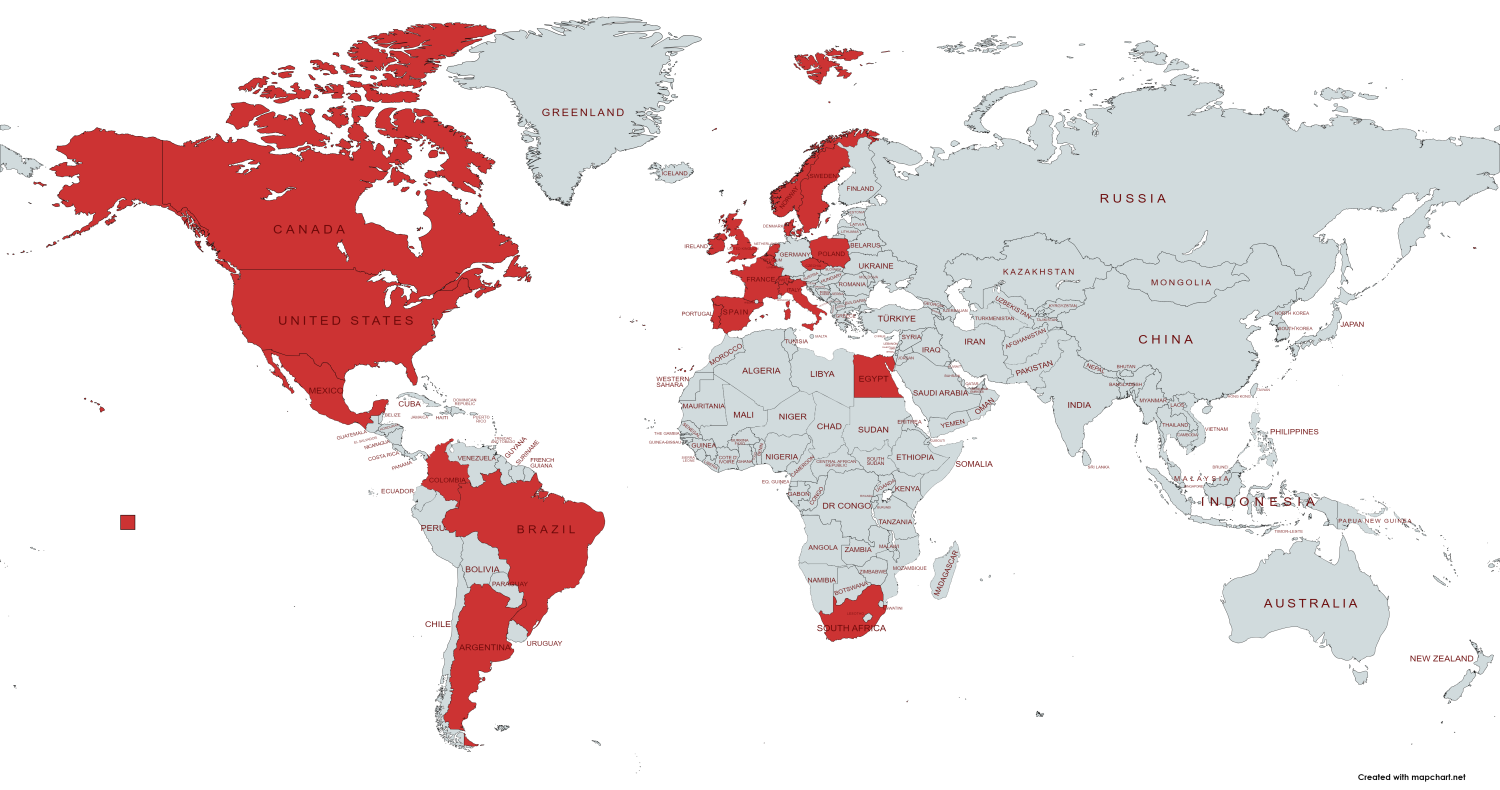
Distribution of REPARES members across the world
[urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 59392 [canonical_url] => [skupina_www] => Array ( ) [url] => /members [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [59393] => stdClass Object ( [nazev] => Antibiotic resistance [seo_title] => Antibiotic resistance [seo_desc] => [autor] => [autor_email] => [obsah] =>Webpage under serious reconstruction! Be back soon!
[urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 59393 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance [sablona] => stdClass Object ( [class] => boxy [html] => [css] => [js] => $(function() { setInterval(function () { $('*[data-countdown]').each(function() { CountDownIt('#'+$(this).attr("id")); }); },1000); setInterval(function () { $('.homebox_slider:not(.stop)').each(function () { slide($(this),true); }); },5000); }); function CountDownIt(selector) { var el=$(selector);foo = new Date; var unixtime = el.attr('data-countdown')*1-parseInt(foo.getTime() / 1000); if(unixtime<0) unixtime=0; var dnu = 1*parseInt(unixtime / (3600*24)); unixtime=unixtime-(dnu*(3600*24)); var hodin = 1*parseInt(unixtime / (3600)); unixtime=unixtime-(hodin*(3600)); var minut = 1*parseInt(unixtime / (60)); unixtime=unixtime-(minut*(60)); if(unixtime<10) {unixtime='0'+unixtime;} if(dnu<10) {unixtime='0'+dnu;} if(hodin<10) {unixtime='0'+hodin;} if(minut<10) {unixtime='0'+minut;} el.html(dnu+':'+hodin+':'+minut+':'+unixtime); } function slide(el,vlevo) { if(el.length<1) return false; var leva=el.find('.content').position().left; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; var cislo=leva/sirka*-1; if(vlevo) { if(cislo+1>pocet) cislo=0; else cislo++; } else { if(cislo==0) cislo=pocet-1; else cislo--; } el.find('.content').animate({'left':-1*cislo*sirka}); el.find('.slider_puntiky a').removeClass('selected'); el.find('.slider_puntiky a.puntik'+cislo).addClass('selected'); return false; } function slideTo(el,cislo) { if(el.length<1) return false; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; if(cislo<0 || cislo>pocet) return false; el.find('.content').animate({'left':-1*cislo*sirka}); el.find('.slider_puntiky a').removeClass('selected'); el.find('.slider_puntiky a.puntik'+cislo).addClass('selected'); return false; } [autonomni] => 1 ) ) [49514] => stdClass Object ( [nazev] => [seo_title] => Media [seo_desc] => [autor] => [autor_email] => [perex] =>Webpage under serious reconstruction! Be back soon!
Newsletter 1 (May 2020)
Newsletter 2 (November 2020)
Newsletter 3 (March 2021)
[ikona] => [obrazek] => [ogobrazek] => [pozadi] => [obsah] => [urlnadstranka] => [iduzel] => 49514 [canonical_url] => [skupina_www] => Array ( ) [url] => /media [sablona] => stdClass Object ( [class] => boxy [html] => [css] => [js] => $(function() { setInterval(function () { $('*[data-countdown]').each(function() { CountDownIt('#'+$(this).attr("id")); }); },1000); setInterval(function () { $('.homebox_slider:not(.stop)').each(function () { slide($(this),true); }); },5000); }); function CountDownIt(selector) { var el=$(selector);foo = new Date; var unixtime = el.attr('data-countdown')*1-parseInt(foo.getTime() / 1000); if(unixtime<0) unixtime=0; var dnu = 1*parseInt(unixtime / (3600*24)); unixtime=unixtime-(dnu*(3600*24)); var hodin = 1*parseInt(unixtime / (3600)); unixtime=unixtime-(hodin*(3600)); var minut = 1*parseInt(unixtime / (60)); unixtime=unixtime-(minut*(60)); if(unixtime<10) {unixtime='0'+unixtime;} if(dnu<10) {unixtime='0'+dnu;} if(hodin<10) {unixtime='0'+hodin;} if(minut<10) {unixtime='0'+minut;} el.html(dnu+':'+hodin+':'+minut+':'+unixtime); } function slide(el,vlevo) { if(el.length<1) return false; var leva=el.find('.content').position().left; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; var cislo=leva/sirka*-1; if(vlevo) { if(cislo+1>pocet) cislo=0; else cislo++; } else { if(cislo==0) cislo=pocet-1; else cislo--; } el.find('.content').animate({'left':-1*cislo*sirka}); el.find('.slider_puntiky a').removeClass('selected'); el.find('.slider_puntiky a.puntik'+cislo).addClass('selected'); return false; } function slideTo(el,cislo) { if(el.length<1) return false; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; if(cislo<0 || cislo>pocet) return false; el.find('.content').animate({'left':-1*cislo*sirka}); el.find('.slider_puntiky a').removeClass('selected'); el.find('.slider_puntiky a.puntik'+cislo).addClass('selected'); return false; } [autonomni] => 1 ) ) [59785] => stdClass Object ( [nazev] => [seo_title] => Public outcomes [seo_desc] => [autor] => [autor_email] => [obsah] =>What happens with antibiotic leftovers that you put in the garbage? And what about the antibiotics that humans and animals are taking?
Are there properly treated? Did they disappear without a problem? Not so much. Have a look at our graphics and learn what is the fate of antibiotics in the environment. If you want to know more, have a look at our Antibiotic resistance page.
Do you think that when you get rid of bacteria in the toilet, it’s over?
Wrong! You send them to a party to make new friends, share experiences, food, and genetic information. Let's see together what is going on at such a party and how it relates to us. Can antibiotics stop all the bacteria? Certainly not all of them, the unstoppable are called superbugs.
Let's see what are these antibiotic-resistant bacteria and what can we do to avoid them...
Combating antibiotic resistance spread
Let's listen to the members of REPARES led by Jan Bartacek talk passionately about why we need to study antibiotic resistance in wastewater treatment plants and the environment. Members from all over Europe talk about how they view antibiotic resistance and what their research in conjunction with REPARES aims to achieve...
Research Platform on Antibiotic REsistance Spread through Wastewater Treatment Plants (REPARES) is an H2020 project dealing with one of the largest threats to human health globally - antibiotic resistance. REPARES brings together both academic and non-academic subjects in combating antibiotic resistance spread in the environment. If your project or research touches on this topic, join REPARES at https://repares.vscht.cz/ and help tackle antibiotic resistance together...
Antibiotic resistance spread through wastewater treatment plants: the race
Immerse yourself in the captivating video by the REPARES Project, as it unveils the intricate web of antibiotic resistance propagation in wastewater treatment plants. Delve into the profound implications of this worldwide health crisis and witness the unwavering determination in the fight against it...
[urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 59785 [canonical_url] => [skupina_www] => Array ( ) [url] => /public-outcomes [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [59396] => stdClass Object ( [nazev] => Contact [seo_title] => Contact [seo_desc] => [autor] => [autor_email] => [obsah] =>Research platform on antibiotic resistance spread through wastewater treatment plants

Jan Bartáček
Project Coordinator, UCT Prague

Lucie Pokorná
Project Administration, Webmaster

Bukola Lois Ojobe
Webmaster
[urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 59396 [canonical_url] => [skupina_www] => Array ( ) [url] => /contact [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [56461] => stdClass Object ( [nazev] => Event registration [seo_title] => Event registration [seo_desc] => [autor] => [autor_email] => [obsah] => [urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 56461 [canonical_url] => [skupina_www] => Array ( ) [url] => /event-registration [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [24134] => stdClass Object ( [obsah] => [iduzel] => 24134 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => [html] => [css] => [js] => [autonomni] => ) ) ) [iduzel] => 49299 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => [html] => [css] => [js] => [autonomni] => ) ) [49298] => stdClass Object ( [obsah] => [poduzel] => stdClass Object ( [49300] => stdClass Object ( [obsah] => [iduzel] => 49300 [canonical_url] => //repares.vscht.cz [skupina_www] => Array ( ) [url] => /[menu-main] [sablona] => stdClass Object ( [class] => api_html [html] => [css] => [js] => [autonomni] => 1 ) ) [49301] => stdClass Object ( [obsah] => [iduzel] => 49301 [canonical_url] => [skupina_www] => Array ( ) [url] => /[menu-top] [sablona] => stdClass Object ( [class] => api_html [html] => [css] => [js] => [autonomni] => 1 ) ) ) [iduzel] => 49298 [canonical_url] => [skupina_www] => Array ( ) [url] => /[en]/49298 [sablona] => stdClass Object ( [class] => api_html [html] => [css] => [js] => [autonomni] => 1 ) ) ) [sablona] => stdClass Object ( [class] => web [html] => [css] => [js] => [autonomni] => 1 ) [api_suffix] => )DATA
stdClass Object
(
[nazev] => Antibiotic resistance
[seo_title] => Antibiotic resistance
[seo_desc] =>
[autor] =>
[autor_email] =>
[autor_no] =>
[obsah] => Webpage under serious reconstruction! Be back soon!
[submenuno] =>
[drobeckyno] =>
[urlnadstranka] =>
[ogobrazek] =>
[pozadi] =>
[newurl_domain] => 'repares.vscht.cz'
[newurl_jazyk] => 'en'
[newurl_akce] => '/antibiotic-resistance'
[newurl_iduzel] => 59393
[newurl_path] => 8549/49296/49297/49299/59393
[newurl_path_link] => Odkaz na newurlCMS
[iduzel] => 59393
[platne_od] => 29.06.2021 13:57:00
[zmeneno_cas] => 29.06.2021 13:57:23.488103
[zmeneno_uzivatel_jmeno] => Lucie Pokorná
[canonical_url] =>
[idvazba] => 70445
[cms_time] => 1751841681
[skupina_www] => Array
(
)
[slovnik] => Array
(
)
[poduzel] => stdClass Object
(
[60332] => stdClass Object
(
[obsah] =>
[poduzel] => stdClass Object
(
[59629] => stdClass Object
(
[nadpis] => Antibiotic Resistance
[odkaz] => https://repares.vscht.cz/antibiotic-resistance/antibiotic-resistance
[text_odkazu] => Antibiotic Resistance
[perex] => - What is AR?
- How does AR spread?
- Information sources
- FAQs
[skupina] =>
[ikona] =>
[velikost] => 3
[pozice_x] => 3
[pozice_y] => 1
[barva_pozadi] => cervena
[countdown] =>
[obrazek_pozadi] => 0004~~cwxSSMqvUMhMzs9TMAEA.jpg
[iduzel] => 59629
[canonical_url] =>
[skupina_www] => Array
(
)
[url] =>
[sablona] => stdClass Object
(
[class] =>
[html] =>
[css] =>
[js] =>
[autonomni] =>
)
)
[59640] => stdClass Object
(
[nadpis] => Wastewater Treatment
[odkaz] => https://repares.vscht.cz/antibiotic-resistance/wastewater-treatment-a-major-barrier-against-ar-spread
[text_odkazu] => Wastewater Treatment: Barrier against AR spread
[perex] => - Technologies
- Legislation
[skupina] =>
[ikona] =>
[velikost] => 3
[pozice_x] => 1
[pozice_y] => 3
[barva_pozadi] => cervena
[countdown] =>
[obrazek_pozadi] => 0005~~Cw8PUUjKr1DITM7PUzAGAA.png
[iduzel] => 59640
[canonical_url] =>
[skupina_www] => Array
(
)
[url] =>
[sablona] => stdClass Object
(
[class] =>
[html] =>
[css] =>
[js] =>
[autonomni] =>
)
)
[59642] => stdClass Object
(
[nadpis] => Exploring wastewater microbiome
[odkaz] => https://repares.vscht.cz/antibiotic-resistance/exploring-wastewater-microbiome
[text_odkazu] => Exploring wastewater microbiome
[perex] => - MiDAS database
- Analytical tools
[skupina] =>
[ikona] =>
[velikost] => 3
[pozice_x] => 2
[pozice_y] => 2
[barva_pozadi] => zelena
[countdown] =>
[obrazek_pozadi] => 0002~~881MLspPyszPVUjKr1DITM7PAwA.png
[iduzel] => 59642
[canonical_url] =>
[skupina_www] => Array
(
)
[url] =>
[sablona] => stdClass Object
(
[class] =>
[html] =>
[css] =>
[js] =>
[autonomni] =>
)
)
)
[iduzel] => 60332
[canonical_url] =>
[skupina_www] => Array
(
)
[url] => /antibiotic-resistance/60332
[sablona] => stdClass Object
(
[class] => stranka_ikona
[html] =>
[css] =>
[js] =>
[autonomni] => 1
)
)
[59718] => stdClass Object
(
[nazev] => Exploring wastewater microbiome
[seo_title] => Exploring wastewater microbiome
[seo_desc] =>
[autor] => Adrian Gorecki, Lukasz Dziewit
[autor_email] =>
[perex] => 
[ikona] =>
[obrazek] =>
[ogobrazek] =>
[pozadi] =>
[obsah] =>
General biodiversity |
|
 |
General biodiversity of the particular microbiome mostly refers to the taxonomic classification of microbes present in a given sample. For this purpose, molecular markers are used. The most common is the 16S rRNA gene, which may be used for specific taxonomic assignments. Data concerning taxonomic diversity may be retrieved from the whole metagenome sequencing projects or specifically targeted metabarcoding approaches. The latter is very common nowadays as it is a cost-effective screening procedure and several analytical tools, e.g., Qiime, are freely available and quite easy to apply. |
Metagenome-based ARGs profiling |
|
| Shotgun metagenomic data give us a broad knowledge of taxonomic structure, metabolic pathways, and other microbiome features. However, sometimes we need precise information, e.g., concerning the presence of ARGs. Retrieving this data usually needs expert knowledge and involves using specific databases, e.g., CARD database. Only by comparing metagenome-retrieved, predicted genes with such databases and applying specific (strict) thresholds one may predict the function of genes and identify them as ARGs. |  |
Gene-based targeted ARGs profiling |
|
 |
For the identification of ARGs, researchers may apply various targeted approaches. These are mostly PCR-based techniques. A literature-based, manually-curated database of PCR primers (LCPDb) for the detection of antibiotic resistance genes that our team members produced contains hundreds of PCR primer pairs designed to amplify various genes conferring resistance to antibiotics. We assigned three parameters for each primer pair: specificity (S), efficacy (E), and taxonomic efficacy (TE). These parameters were evaluated using a novel bioinformatic tool – UniPriVal – used to validate each primer pair against various reference databases. Then, we ranked primer pairs specific for each gene based on their model success metric (MSM) value. Despite this limitation, the internal validation system of the LCPDb application enables the quantified ranking of PCR primer pairs, which assists the selection of the best primers for each application. |
Functional (meta)genomics |
|
| In the last decade, the costs of DNA sequencing have significantly decreased, and the time of such analysis was significantly shortened. A huge number of available DNA sequences (including genomes and metagenomes) resulted in the development of various tools for (meta)genomes annotation. One can perform automatic annotation with various pipelines, however manual curation of the results of such annotation is still necessary to obtain reliable and strictly validated data. Unfortunately, due to its time-consuming nature, manual annotation is now rarely used. Several bioinformatics tools can be used for manual annotation of bacteria genomes. Here we present our novel invention – MAISEN. MAISEN is a free, web-based tool designed to accelerate manual annotation using a simple interface and precomputed alignments for each predicted feature. It was designed to be available for every user (also non-experts). |  |
Identification of gene variants |
|
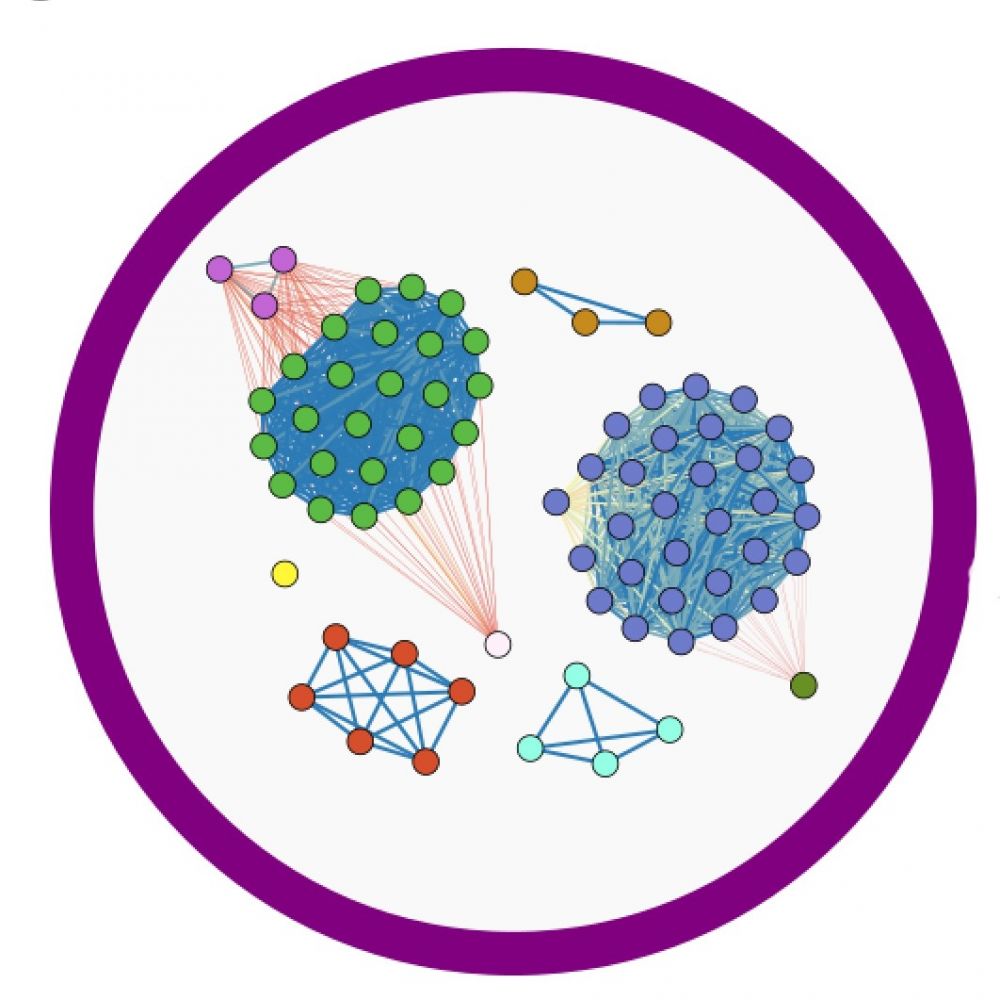 |
An increasing number of novel ARG variants require tools that help to detect all of them. Combining clustering models and multiple sequence alignments, a novel tool was designed to provide users with primer pairs that cover all possible variants of the particular ARG. The ConPrimise designs the set of primers specific to a particular gene using a consensus sequence calculated from all known variants of the particular ARG after multiple sequence alignments. From the developed set of primer pairs, with the usage of UniPriVal, we can select primers with maximal efficacy statistics to prevent skipping any variant during the PCR screening. |
In REPARES, we developed a dedicated database of full genomes of antibiotic-resistant bacteria found at various WWTPs. The database is embedded into the MiDAS field guide funded and operated by Aalborg University.
[iduzel] => 59833 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => infobox [html] => [css] => [js] => [autonomni] => 0 ) ) [73186] => stdClass Object ( [nazev] => REPARES interlaboratory study (ring test) [barva_pozadi] => cervena [uslideru] => false [text] =>qPCR quantification of antibiotic resistance genes and other related genes in DNA isolated from sewage sludge and wastewater accessible through ZENODO repository
[iduzel] => 73186 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => infobox [html] => [css] => [js] => [autonomni] => 0 ) ) ) [iduzel] => 59718 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance/exploring-wastewater-microbiome [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [49345] => stdClass Object ( [obsah] => [poduzel] => stdClass Object ( [49346] => stdClass Object ( [nadpis] => [popis] => [platne_od] => 19.09.2019 [platne_do] => 19.09.2030 [odkaz] => [text_odkazu] => [obrazek_pozadi] => 0003~~C_bxdHENUshLVAh3dVIoSS0uAQA.jpg [barva_textu] => [iduzel] => 49346 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => slider [html] => [css] => [js] => [autonomni] => 0 ) ) ) [iduzel] => 49345 [canonical_url] => _clone_ [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => slider [html] => [css] => [js] => [autonomni] => 0 ) ) [59717] => stdClass Object ( [nazev] => Antibiotic Resistance [seo_title] => Antibiotic Resistance [seo_desc] => [autor] => Ivone vaz Moreira, Celia M. Manaia [autor_email] => [perex] =>What is antibiotic resistance?
Antibiotic resistance is the capacity of bacteria to survive or proliferate in presence of antibiotics. Many bacteria in nature have this capacity. Most of the antibiotics commercially available nowadays are derivatives of natural compounds produced by bacteria and/or fungi.
More information
In nature, microorganisms may use antibiotics as a defense mechanism, by inhibiting the growth of competitors, but antibiotics may have other roles.
Before the introduction of antibiotics in the 1940s as therapeutic agents antibiotics were only found in nature. After that period, the increasing uses in human medicine for bacterial infection therapy, and in veterinary for growth-promotion or infection prophylaxis contributed to a widespread occurrence of antibiotics and, with them, antibiotic-resistant bacteria. Indeed, the increasing use of antibiotics, and other substances with antimicrobial activity, pushed antibiotic-resistant bacteria to reach new habitats, waters, soil, wildlife...
How can bacteria become resistant to antibiotics?
Antibiotic resistance may be intrinsic or acquired. Intrinsic resistance is characteristic of a bacterial species or genus, it is present in most or all members of that group and probably it was inherited from their evolutionary ancestors.
More information
Hence, it is part of the genome of most members of the group, it will be favored by antibiotic selective pressure, and is always transmitted vertically, i.e. from parent cells to the progeny. Intrinsic antibiotic resistance can be associated with structural or functional characteristics of a species, for example, due to the absence of a peptidoglycan cell wall or the existence of an external membrane, typical of Gram-negative bacteria.
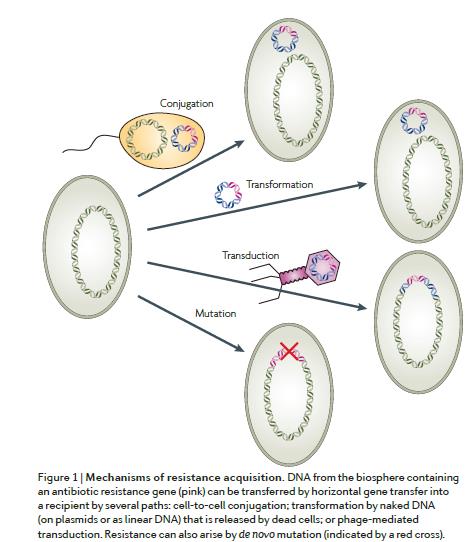
Andersson and Hughes, 2010
The type of antibiotic resistance that is considered a major disseminating threat is, however, acquired. This means that is a property that develops in a bacterial cell that was once susceptible to a given antibiotic. This process may occur due to point (or gene) mutations or, more frequently, through horizontal gene transfer (HGT). HGT is a major driver of bacterial evolution and consists in the transfer of genetic material from a donor to a recipient cell, in general, it requires that both share the same space, but not necessarily the same species. HGT can occur by transformation, consisting on the uptake of naked DNA (on plasmids or as linear DNA), for example, released by dead cells; transduction, mediated by bacteriophages (the virus of bacteria); or conjugation, involving cell-to-cell contact through a pilus. In general, HGT processes are potentiated by genetic elements that facilitate the mobilization and integration of exogenous DNA, either between cells or between chromosomal DNA, and extrachromosomal genetic elements and vice versa. Examples of these genetic elements are plasmids, transposons, and integrons, in which many of the known antibiotic resistance genes are inserted.
Mechanisms of antibiotic resistance
The mechanisms of antibiotic resistance comprise i) modifications of the antibiotic, ii) prevention of antibiotic action (by decreasing penetration or actively extruding the antimicrobial compound), and iii) changes and/or bypass of target sites.
More information
More detail about these mechanisms is available in several publications, as for example:
- Munita&Arias, 2016; https://onlinelibrary.wiley.com/doi/abs/10.1128/9781555819286.ch17.
- Yelin&Kishony, 2018;https://www.cell.com/cell/pdf/S0092-8674(18)30162-4.pdf
How antibiotic resistance does spread?
Human activities lead to the continuous discharge of antibiotic-resistant bacteria and genes to the environment, which accumulates and spread over different environmental compartments. Although the natural antibiotic resistome (collection of antibiotic resistance genes naturally occurring in the environment) was the origin of the antibiotic-resistance genes that are currently of major concern at the clinical level, it is the contaminant resistome (collection of antibiotic resistance genes resulting from human activities) that represents the major risk of transmission to humans.
More information
The high loads of contaminant antibiotic-resistant bacteria continuously discharged to the environment, and the fact that the contaminant resistome comprises bacteria and genes that coevolved with animals and humans, hence with higher chances of colonizing humans and animals, are major reasons to give the contaminant resistome particular attention. However, only some bacteria constituting the contaminant resistome may have the capacity to colonize humans (the vectors), being pivotal for assessing the risks of transmission of antibiotic resistance from the environment to humans. Other bacteria of the contaminant resistome that, for some reason, cannot colonize humans (the carriers), also have a role in the spread of antibiotic-resistance genes in the environment and may contribute to the enrichment of vectors of antibiotic-resistance genes.
[ikona] => [obrazek] => 0001~~88rPzFMIcg1wDHINVtAw0gQA.png [ogobrazek] => [pozadi] => [obsah] =>
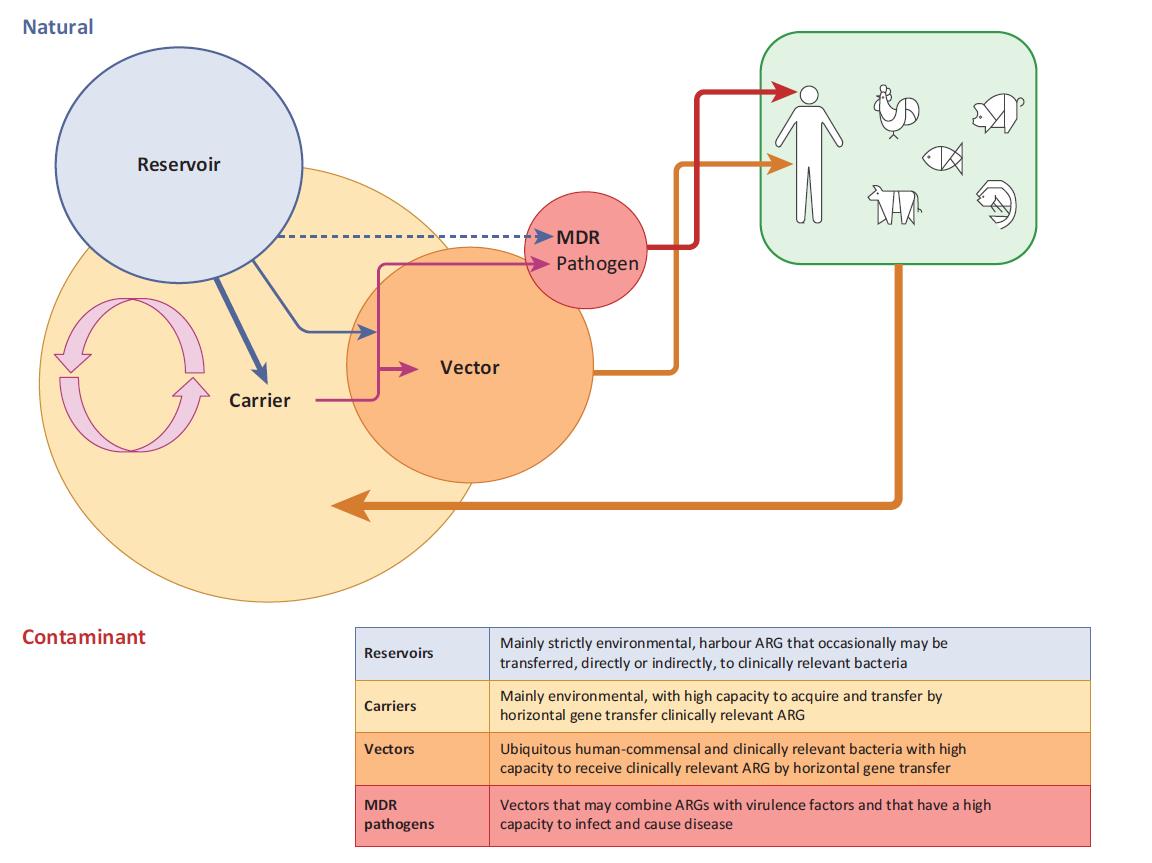 Manaia 2017
Manaia 2017
References:
- Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., & Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nature reviews microbiology, 13(1), 42-51.
- D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W., Schwarz, C., ... & Wright, G. D. (2011). Antibiotic resistance is ancient. Nature, 477(7365), 457-461.
- Andersson, D. I., & Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance?. Nature Reviews Microbiology, 8(4), 260-271.
- Munita, J. M., & Arias, C. A. (2016). Mechanisms of antibiotic resistance. Virulence mechanisms of bacterial pathogens, 481-511.
- Yelin, I., & Kishony, R. (2018). Antibiotic resistance. Cell, 172(5), 1136-1136.
- Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends in microbiology, 25(3), 173-181.
[urlnadstranka] => [poduzel] => stdClass Object ( [59837] => stdClass Object ( [nazev] => Information sources [barva_pozadi] => cervena [uslideru] => false [text] => [iduzel] => 59837 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => infobox [html] => [css] => [js] => [autonomni] => 0 ) ) [59838] => stdClass Object ( [nazev] => FAQs [barva_pozadi] => cervena [uslideru] => false [text] => [iduzel] => 59838 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => infobox [html] => [css] => [js] => [autonomni] => 0 ) ) [59757] => stdClass Object ( [nazev] => Information sources [seo_title] => Information sources [seo_desc] => [autor] => [autor_email] => [obsah] =>
Antibiotic consumption
Human antimicrobial consumption database (ESAC-Net) from the European Centre for Disease Prevention and Control (ECDC)
ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food‐producing animals
European Surveillance of Veterinary Antimicrobial Consumption (ESVAC) from the European Medicines Agency (EMA)
Antimicrobial resistance
Antimicrobial resistance in Europe - Data visualization tool from the European Centre for Disease Prevention and Control (ECDC)
ECDC Surveillance Atlas of Antimicrobial resistance
European Committee on Antimicrobial Susceptibility Testing (EUCAST)
ResistanceMap - an interactive collection of charts and maps that summarize national and subnational data on antimicrobial use and resistance worldwide, by The Center for Disease Dynamics, Economics & Policy (CDDEP)
Antibiotic resistance genes
The Comprehensive Antibiotic Resistance Database (CARD)
ARGs-OSP: Antibiotic-Resistant Genes Online Searching Platform
[urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 59757 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance/antibiotic-resistance/information-sources [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [59839] => stdClass Object ( [nazev] => FAQs [seo_title] => FAQs [seo_desc] => [autor] => [autor_email] => [obsah] =>What is antibiotic resistance?
Antibiotic resistance refers to bacteria and it is particularly important when these are pathogens. In such a case, antibiotic therapy will fail. Hence, “antibiotic resistance” is a human- and animal health issue. However, antibiotic resistance has been regarded as biological pollution that can occur and spread in the environment, and be returned to humans and animals.
What does it mean when bacteria are resistant to antibiotics?
It means that they will be able to proliferate in the presence of the antibiotic, meaning that the therapeutical agent is useless. More info...
What are “Superbugs”?
“Superbugs” is a non-scientific word that refers to bacteria that have the capacity to proliferate in presence of different types of antibiotics, leaving the doctors very limited or no alternatives to treat bacterial infections. In technical terms, a superbug refers to a multidrug-resistant bacteria, defined as those able to resist antibiotics that belong to at least three different classes. In extreme cases, a superbug may be resistant to all the antibiotics available.
How does antibiotic resistance happen?
It can be an intrinsic property of bacteria, related to their structure or metabolism. In other cases it can be acquired when bacteria gain new genetic information, which normally occurs through horizontal gene transfer, meaning that it is the passage of genetic material between contemporaneous cells. This contrasts with the vertical gene transfer corresponding to the transmission of genetic material from ancestors to progeny. Antibiotic resistance may occur by different mechanisms. See also Mechanisms of antibiotic resistance
Is antibiotic resistance an important issue?
Antibiotic resistance can affect anyone, of any age and in any country. In Europe, where good health and sanitation conditions are in place, can kill about 30 000 people per year. Moreover, antibiotic resistance leads to increased medical costs, prolonged hospital stays failure of medical interventions, and higher mortality. For all these reasons it has become a subject of major concern for public health authorities.
What is the single most significant factor driving increasing rates of antibiotic resistance?
The factor pointed as most significant is the use of antibiotics. For that reason many awareness campaigns are being done to reduce the prescription and use of antibiotics, mainly to avoid the profilatic use and save antibiotics to be used just when they are really needed. Once disseminated in the human and animal microbiomes and in the environment, there are multiple factors that drive the increase of antibiotic resistance, such as pollution, poor sanitation conditions, or deficient healthcare assistance.
Can the same bacteria be resistant to several antibiotics?
Yes, these are the so-called superbugs that can resist to several antibiotics. This capacity is acquired through the gain of successive resistance pieces, scientifically designated as antibiotic resistance genes. See also the FAQ “What are superbugs?”
Why is it important to take antibiotics exactly as prescribed?
The prescription should be always made based on the identification of the infectious bacteria as well as the antibiotic susceptibility profile. Once this information is available, the dose prescribed should be required to efficiently eliminate the infectious agent. If treatment is halted before the due time, there is the risk that only part of the infectious bacteria was killed and these will multiply making the person sick again. It is possible that bacteria that were not eliminated were those with the highest level of resistance, making the second infection even more difficult to treat. For the same reasons, antibiotic intakes doses should not be skipped.
Very important: antibiotics must always be taken after medical prescription, and based on a microbiological diagnostic to determine the cause of the disease. The wrong medicine will delay the treatment.
More info: https://www.fda.gov/consumers/consumer-updates/combating-antibiotic-resistance
What are the main causes of antibiotic resistance?
In addition to the excessive, and not always appropriate, use of antibiotics other causes may be contributing to the increase of antibiotic resistance. For example, 1) in the human body, a poor or unbalanced microbiome will give the antibiotic-resistant bacteria opportunity to proliferate; 2) in food-animal production the use of antibiotics favour the overproliferation of antibiotic-resistant bacteria; 3) in the environment, eutrophic systems with high load of nutrients and pollution will favor the proliferation of antibiotic resistant bacteria.
These three examples show the importance of the One-Health concept on the dissemination of antibiotic resistance, assuming the links on the occurrence of antibiotic resistance in humans, animals, and the environment.
How can antimicrobial resistance be measured/valued?
The method most commonly used to measure the susceptibility of a bacterium to different concentrations of antibiotic in either broth or agar culture media or on paper discs. The lowest antibiotic concentration that inhibits bacterial growth is the minimum inhibitory concentration (MIC).
The application of molecular biology techniques, as quantitative PCR, allows the quantification of genetic determinants related to antibiotic resistance (antibiotic resistance genes).
More information:
- Davison, H. C., Woolhouse, M. E., & Low, J. C. (2000). What is antibiotic resistance and how can we measure it?. Trends in microbiology, 8(12), 554-559.
- Manaia, Célia M., Jaqueline Rocha, Nazareno Scaccia, Roberto Marano, Elena Radu, Francesco Biancullo, Francisco Cerqueira et al. "Antibiotic resistance in wastewater treatment plants: tackling the black box." Environment International 115 (2018): 312-324.
What can I do to help prevent antibiotic resistance?
- Take the antibiotics as prescribed.
- Do not skip doses.
- Do not save antibiotics.
- Do not take antibiotics prescribed for someone else.
- Talk with your health care professional.
- All drugs have side effects.
- Take care of environmental sustainability, avoiding the emission of pollutants, contributing to the preservation of natural resources such as soil and water and, protecting biodiversity.
More info can be found here.
Does antibiotic resistance go away?
Resistance does not go away.
Most acquired antibiotic resistance mechanisms are not associated with a fitness cost for the bacteria, meaning that the presence of that resistance type in the bacterial cells does not affect its survival or proliferation rate. The practical implication of this is that once acquired, a resistance type will not be lost, even when the antibiotic is no longer used.
Andersson, D. I., & Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance?. Nature Reviews Microbiology, 8(4), 260-271.
[urlnadstranka] => [ogobrazek] => [pozadi] => [iduzel] => 59839 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance/antibiotic-resistance/faqs [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) ) [iduzel] => 59717 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance/antibiotic-resistance [sablona] => stdClass Object ( [class] => stranka_obrazek_vertical [html] => [css] => [js] => [autonomni] => 1 ) ) [59715] => stdClass Object ( [nazev] => [seo_title] => Wastewater Treatment: A major barrier against AR spread [seo_desc] => [autor] => [autor_email] => [obsah] =>The vast majority of antibiotic-resistant bacteria (ARB) develop in the bodies of antibiotic-treated patients (human or animal) and are later excreted in feces and urine into sewage. Municipal, hospital, industrial, and (sometimes) agricultural wastewaters are collected and treated in wastewater treatment plants (WWTPs), where the presence of diverse selection pressures together with a highly concentrated consortium of pathogenic microbes create favorable conditions for further transfer of antibiotic resistance genes (ARGs) and proliferation of ARB. The rapid emergence of antibiotic-resistant pathogens of clinical and veterinary significance over the past 80 years has redefined the role of WWTPs as a focal point in the fight against antimicrobial resistance (AMR). Wastewater treatment is a double-edged sword that can act as either a pathway for AMR spread or as a barrier to reduce the environmental release of anthropogenic AMR.
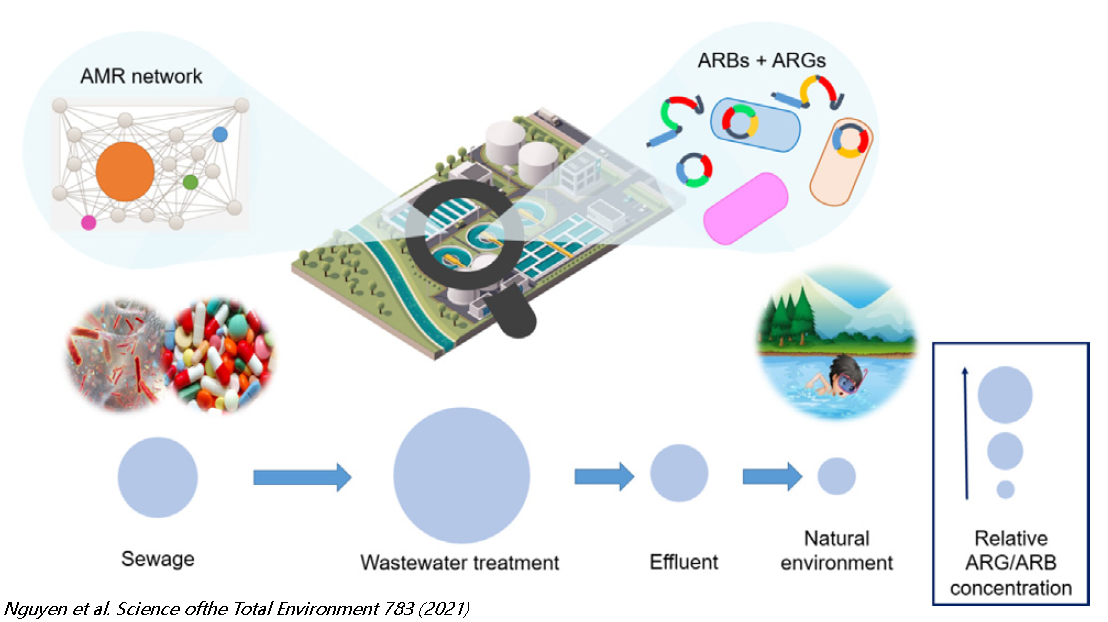
If properly designed and operated, current wastewater treatment processes such as activated sludge process (AS), membrane bioreactors (MBRs), biofilters (BFs), or constructed wetlands (CWs) can efficiently stop both ARGs and ARB from entering WWTP's effluent under the condition that bacteria are sufficiently retained inside the WWTP. Therefore, technologies with highly efficient biomass retention (e.g. MBR) are most efficient in ARG/ARB removal. Even then, extracellular DNA (or mobile genetic elements - MGE) can escape to recipents (rivers, lakes, oceans, or groundwater). To fully remove ArRGs and ARB from WWTPs' effluents, tertiary treatment with advanced techniques such as UV irradiation, ozonation, Fenton reaction, or photocatalysis is required.
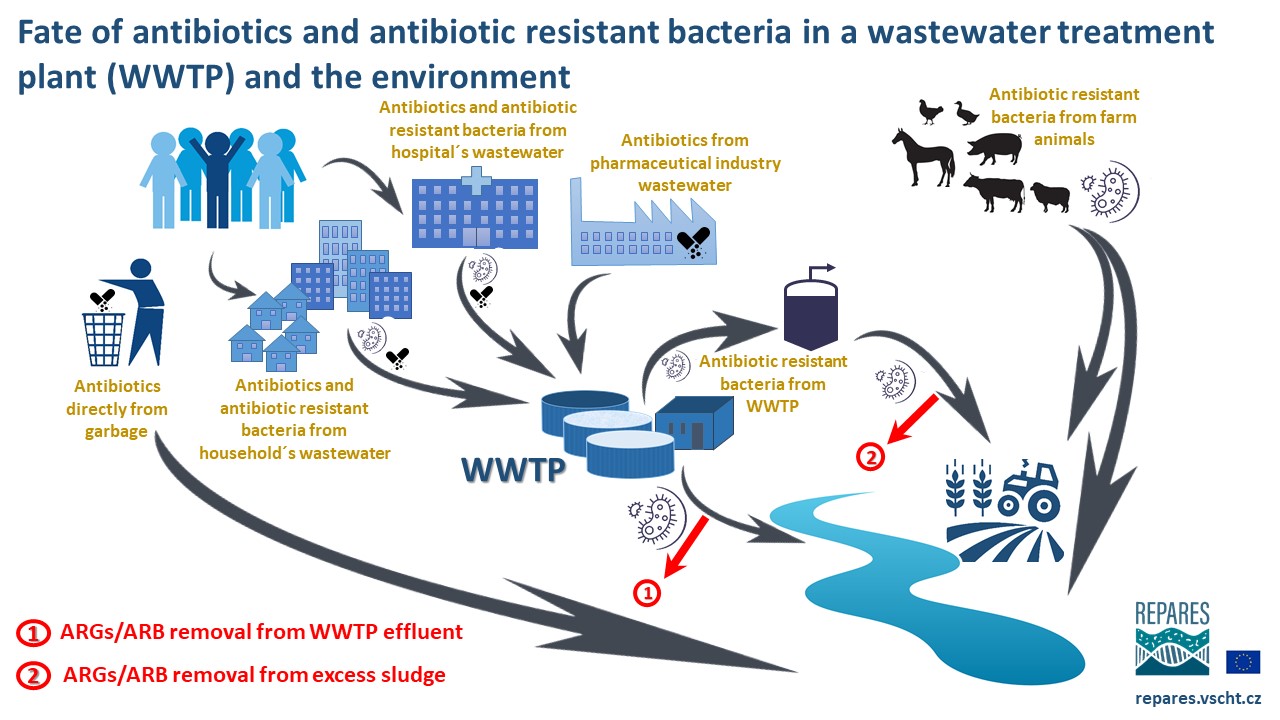
Furthermore, ARGs and ARB accumulate in the excess activated sludge (microbial biomass growing as a result of biological wastewater treatment) and must be destroyed before its further utilization. Most of the waste sludge is first subjected to anaerobic digestion and then utilized for energy production (incineration, gasification, etc.) or in agriculture as fertilizer. In the latter case, sludge hygienization such as pasteurization, drying, liming or thermal hydrolysis, may help to remove ARGs/ARB.
[urlnadstranka] => [ogobrazek] => [pozadi] => [poduzel] => stdClass Object ( [49345] => stdClass Object ( [obsah] => [iduzel] => 49345 [canonical_url] => _clone_ [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => slider [html] => [css] => [js] => [autonomni] => 0 ) ) [60333] => stdClass Object ( [nazev] => [barva_pozadi] => cervena [uslideru] => false [text] => [iduzel] => 60333 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => infobox [html] => [css] => [js] => [autonomni] => 0 ) ) [60334] => stdClass Object ( [nazev] => [barva_pozadi] => cervena [uslideru] => false [text] => [iduzel] => 60334 [canonical_url] => [skupina_www] => Array ( ) [url] => [sablona] => stdClass Object ( [class] => infobox [html] => [css] => [js] => [autonomni] => 0 ) ) ) [iduzel] => 59715 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance/wastewater-treatment-a-major-barrier-against-ar-spread [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) [59798] => stdClass Object ( [nazev] => Interlaboratory Study [seo_title] => Interlaboratory Study [seo_desc] => [autor] => Sabina Purkrtova [autor_email] => [perex] =>The methods for the quantification of ARGs enable quantifying ARGs amount in different parts of wastewater treatment plants (WWTPs), including inputs and outputs. This data allows to survey the fate of ARGs in WWTPs, to study the effect of single processes in WWTP on ARGs abundance, and last but not least to quantify the applied load of ARGs carried by WWTP outputs to the environments.
[ikona] => [obrazek] => [ogobrazek] => [pozadi] => [obsah] => The quantitative-based PCR methods (qPCR) belong to the most often used methods for quantifying ARGs, widely used in scientific studies. Despite this fact, the standard internationally validated or recommended methodology for qPCR quantification in WWTP samples has not yet been established. The quantitative standard methodology (“golden standard”) would cover not only the description of the single procedure steps but also the evaluation of test characteristics, for example, the selectivity (inclusivity/exclusivity), lowest validated level with satisfactory precision, repeatability, reproducibility and the uncertainty of the method.
The quantitative-based PCR methods (qPCR) belong to the most often used methods for quantifying ARGs, widely used in scientific studies. Despite this fact, the standard internationally validated or recommended methodology for qPCR quantification in WWTP samples has not yet been established. The quantitative standard methodology (“golden standard”) would cover not only the description of the single procedure steps but also the evaluation of test characteristics, for example, the selectivity (inclusivity/exclusivity), lowest validated level with satisfactory precision, repeatability, reproducibility and the uncertainty of the method.
In REPARES Ring test interlaboratory studies, we work on establishing such qPCR-based standard protocol and on evaluating the effect of single steps on its result. These single steps cover the isolation of DNA and absolute qPCR quantification based on DNA intercalating dyes and using gBlocks.
At this moment, these interlaboratory studies are four sides, conducted by UCP Porto, UCT Prague, TU Delft, and Wetsus. The University of Warsaw ensures the bioinformatics support for these studies.
In the first run in 2020, we analyzed qPCR quantification of four different genes in activated sludges. All laboratories analyzed as the sample of activated sludges, as the sample DNA, isolated from it. The results imply that qPCR analyses have more extent effect on these ARGs quantification than performing the isolation of DNA.
In the second run, planned for September 2021, we will test these qPCR protocols for samples of anaerobic sludges and treated WWTP effluent and some other genes. The important part of the investigation will also direct to discover and quantify the impact of single qPCR steps (e.g., primers design, used master mixes, and chemicals).
We want to invite also other laboratories to join our interlaboratory studies.
We are looking for cooperating laboratories quantifying ARGs. It enables us to fortify these protocols verification (by testing our protocol if you perform qPCR quantification too) and/or to compare these protocols to other ARGs quantification methods (by comparing results if you perform different qPCR protocols or any other DNA based quantification methods).
If you are interested in joining us, please write to repares@vscht.cz subject: RING TEST. We will find a way how to cooperate.
[urlnadstranka] => [poduzel] => Array ( ) [iduzel] => 59798 [canonical_url] => [skupina_www] => Array ( ) [url] => /antibiotic-resistance/interlaboratory-study [sablona] => stdClass Object ( [class] => stranka [html] => [css] => [js] => [autonomni] => 1 ) ) ) [sablona] => stdClass Object ( [class] => boxy [html] => [css] => [js] => $(function() { setInterval(function () { $('*[data-countdown]').each(function() { CountDownIt('#'+$(this).attr("id")); }); },1000); setInterval(function () { $('.homebox_slider:not(.stop)').each(function () { slide($(this),true); }); },5000); }); function CountDownIt(selector) { var el=$(selector);foo = new Date; var unixtime = el.attr('data-countdown')*1-parseInt(foo.getTime() / 1000); if(unixtime<0) unixtime=0; var dnu = 1*parseInt(unixtime / (3600*24)); unixtime=unixtime-(dnu*(3600*24)); var hodin = 1*parseInt(unixtime / (3600)); unixtime=unixtime-(hodin*(3600)); var minut = 1*parseInt(unixtime / (60)); unixtime=unixtime-(minut*(60)); if(unixtime<10) {unixtime='0'+unixtime;} if(dnu<10) {unixtime='0'+dnu;} if(hodin<10) {unixtime='0'+hodin;} if(minut<10) {unixtime='0'+minut;} el.html(dnu+':'+hodin+':'+minut+':'+unixtime); } function slide(el,vlevo) { if(el.length<1) return false; var leva=el.find('.content').position().left; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; var cislo=leva/sirka*-1; if(vlevo) { if(cislo+1>pocet) cislo=0; else cislo++; } else { if(cislo==0) cislo=pocet-1; else cislo--; } el.find('.content').animate({'left':-1*cislo*sirka}); el.find('.slider_puntiky a').removeClass('selected'); el.find('.slider_puntiky a.puntik'+cislo).addClass('selected'); return false; } function slideTo(el,cislo) { if(el.length<1) return false; var sirka=el.width(); var pocet=el.find('.content .homebox').length-1; if(cislo<0 || cislo>pocet) return false; el.find('.content').animate({'left':-1*cislo*sirka}); el.find('.slider_puntiky a').removeClass('selected'); el.find('.slider_puntiky a.puntik'+cislo).addClass('selected'); return false; } [autonomni] => 1 ) [api_suffix] => )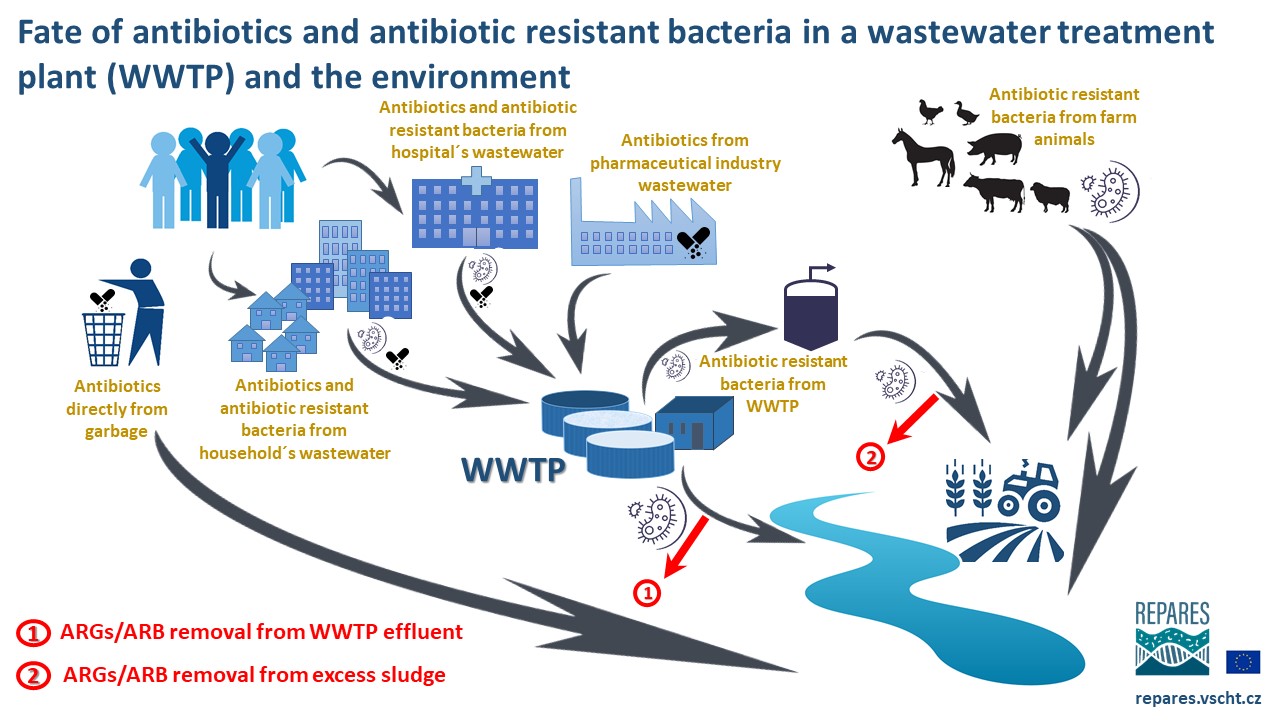

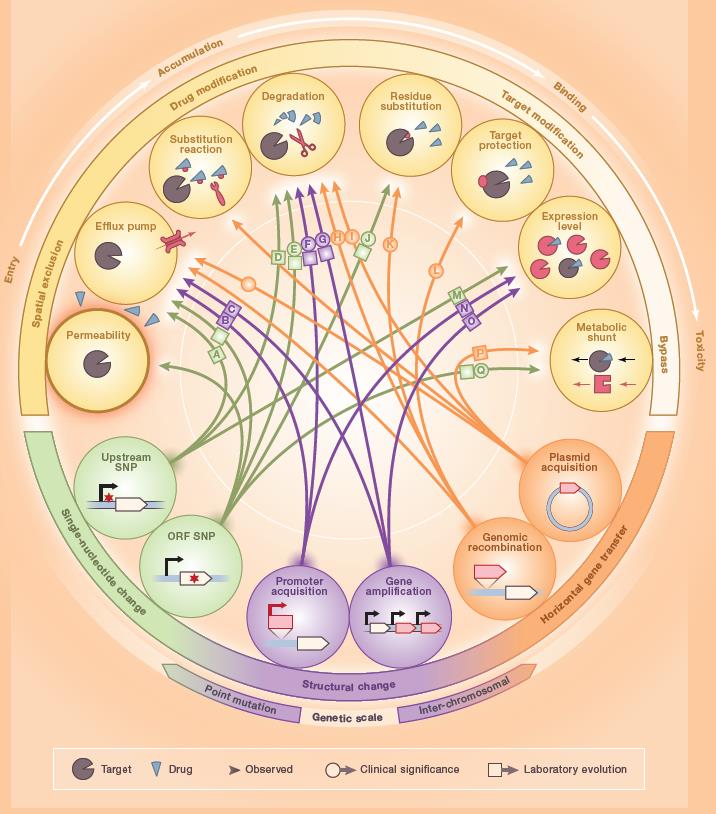 Yalin and Kishony 2018
Yalin and Kishony 2018


