
What is antibiotic resistance?
Antibiotic resistance is the capacity of bacteria to survive or proliferate in presence of antibiotics. Many bacteria in nature have this capacity. Most of the antibiotics commercially available nowadays are derivatives of natural compounds produced by bacteria and/or fungi.
More information
In nature, microorganisms may use antibiotics as a defense mechanism, by inhibiting the growth of competitors, but antibiotics may have other roles.
Before the introduction of antibiotics in the 1940s as therapeutic agents antibiotics were only found in nature. After that period, the increasing uses in human medicine for bacterial infection therapy, and in veterinary for growth-promotion or infection prophylaxis contributed to a widespread occurrence of antibiotics and, with them, antibiotic-resistant bacteria. Indeed, the increasing use of antibiotics, and other substances with antimicrobial activity, pushed antibiotic-resistant bacteria to reach new habitats, waters, soil, wildlife...
How can bacteria become resistant to antibiotics?
Antibiotic resistance may be intrinsic or acquired. Intrinsic resistance is characteristic of a bacterial species or genus, it is present in most or all members of that group and probably it was inherited from their evolutionary ancestors.
More information
Hence, it is part of the genome of most members of the group, it will be favored by antibiotic selective pressure, and is always transmitted vertically, i.e. from parent cells to the progeny. Intrinsic antibiotic resistance can be associated with structural or functional characteristics of a species, for example, due to the absence of a peptidoglycan cell wall or the existence of an external membrane, typical of Gram-negative bacteria.
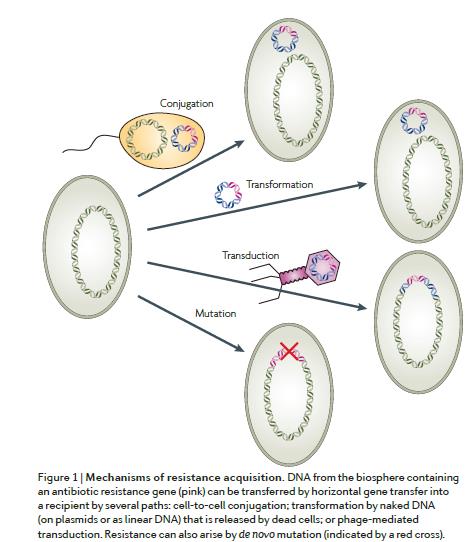
Andersson and Hughes, 2010
The type of antibiotic resistance that is considered a major disseminating threat is, however, acquired. This means that is a property that develops in a bacterial cell that was once susceptible to a given antibiotic. This process may occur due to point (or gene) mutations or, more frequently, through horizontal gene transfer (HGT). HGT is a major driver of bacterial evolution and consists in the transfer of genetic material from a donor to a recipient cell, in general, it requires that both share the same space, but not necessarily the same species. HGT can occur by transformation, consisting on the uptake of naked DNA (on plasmids or as linear DNA), for example, released by dead cells; transduction, mediated by bacteriophages (the virus of bacteria); or conjugation, involving cell-to-cell contact through a pilus. In general, HGT processes are potentiated by genetic elements that facilitate the mobilization and integration of exogenous DNA, either between cells or between chromosomal DNA, and extrachromosomal genetic elements and vice versa. Examples of these genetic elements are plasmids, transposons, and integrons, in which many of the known antibiotic resistance genes are inserted.
Mechanisms of antibiotic resistance
The mechanisms of antibiotic resistance comprise i) modifications of the antibiotic, ii) prevention of antibiotic action (by decreasing penetration or actively extruding the antimicrobial compound), and iii) changes and/or bypass of target sites.
More information
More detail about these mechanisms is available in several publications, as for example:
- Munita&Arias, 2016; https://onlinelibrary.wiley.com/doi/abs/10.1128/9781555819286.ch17.
- Yelin&Kishony, 2018;https://www.cell.com/cell/pdf/S0092-8674(18)30162-4.pdf
How antibiotic resistance does spread?
Human activities lead to the continuous discharge of antibiotic-resistant bacteria and genes to the environment, which accumulates and spread over different environmental compartments. Although the natural antibiotic resistome (collection of antibiotic resistance genes naturally occurring in the environment) was the origin of the antibiotic-resistance genes that are currently of major concern at the clinical level, it is the contaminant resistome (collection of antibiotic resistance genes resulting from human activities) that represents the major risk of transmission to humans.
More information
The high loads of contaminant antibiotic-resistant bacteria continuously discharged to the environment, and the fact that the contaminant resistome comprises bacteria and genes that coevolved with animals and humans, hence with higher chances of colonizing humans and animals, are major reasons to give the contaminant resistome particular attention. However, only some bacteria constituting the contaminant resistome may have the capacity to colonize humans (the vectors), being pivotal for assessing the risks of transmission of antibiotic resistance from the environment to humans. Other bacteria of the contaminant resistome that, for some reason, cannot colonize humans (the carriers), also have a role in the spread of antibiotic-resistance genes in the environment and may contribute to the enrichment of vectors of antibiotic-resistance genes.
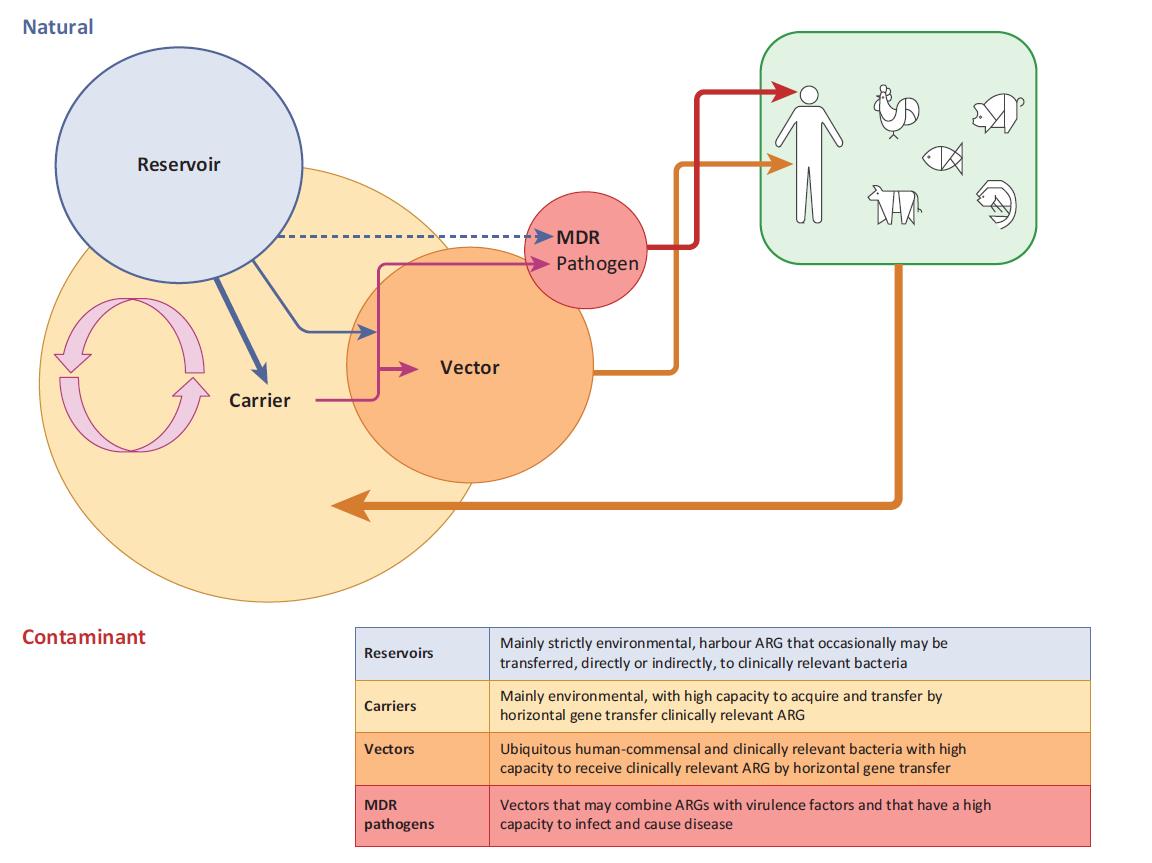 Manaia 2017
Manaia 2017
References:
- Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., & Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nature reviews microbiology, 13(1), 42-51.
- D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W., Schwarz, C., ... & Wright, G. D. (2011). Antibiotic resistance is ancient. Nature, 477(7365), 457-461.
- Andersson, D. I., & Hughes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance?. Nature Reviews Microbiology, 8(4), 260-271.
- Munita, J. M., & Arias, C. A. (2016). Mechanisms of antibiotic resistance. Virulence mechanisms of bacterial pathogens, 481-511.
- Yelin, I., & Kishony, R. (2018). Antibiotic resistance. Cell, 172(5), 1136-1136.
- Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends in microbiology, 25(3), 173-181.

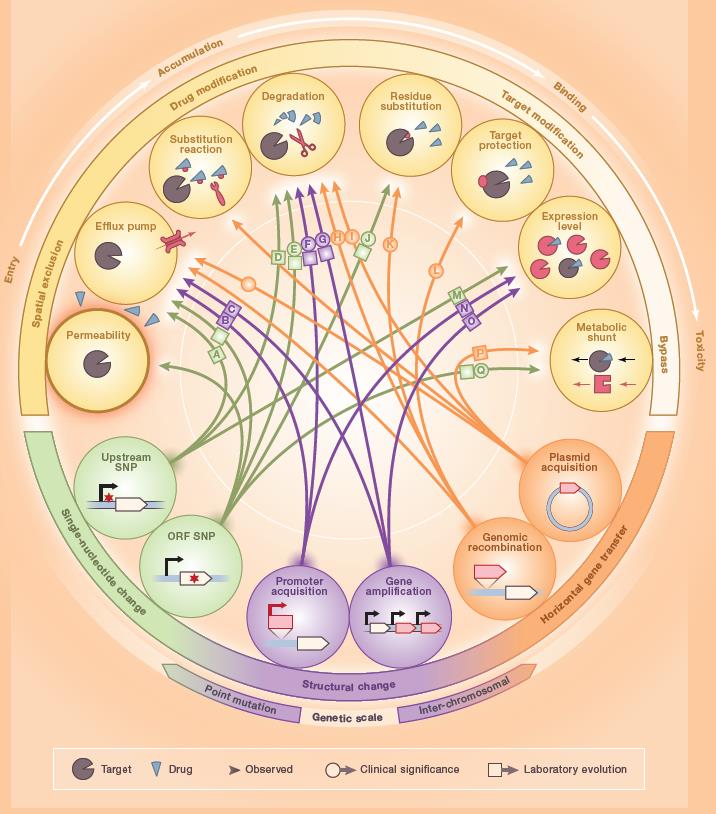 Yalin and Kishony 2018
Yalin and Kishony 2018
